import act
import numpy as np
import xarray as xr
import matplotlib.pyplot as plt
import matplotlib.colors as colors# Set your username and token here!
username = 'iregmisid'
#token =
# Set the datastream and start/enddates
datastream = 'bnfecorsfS20.b1'
startdate = '2025-04-01T00:00:00'
enddate = '2025-04-30T23:59:59'
# Use ACT to easily download the data. Watch for the data citation! Show some support
# for ARM's instrument experts and cite their data if you use it in a publication
result_ecor = act.discovery.download_arm_data(username, token, datastream, startdate, enddate)
[DOWNLOADING] bnfecorsfS20.b1.20250417.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250430.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250411.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250405.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250409.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250424.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250418.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250423.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250420.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250416.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250407.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250404.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250406.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250414.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250410.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250419.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250401.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250403.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250428.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250412.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250427.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250426.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250422.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250402.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250421.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250415.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250408.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250425.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250429.000000.nc
[DOWNLOADING] bnfecorsfS20.b1.20250413.000000.nc
If you use these data to prepare a publication, please cite:
Sullivan, R., Cook, D., Shi, Y., Keeler, E., & Pal, S. Eddy Correlation Flux
Measurement System (ECORSF), 2025-04-01 to 2025-04-30, Bankhead National Forest,
AL, USA; Long-term Mobile Facility (BNF), Bankhead National Forest, AL,
Supplemental facility at Courtland (S20). Atmospheric Radiation Measurement
(ARM) User Facility. https://doi.org/10.5439/1494128
ds_ecor = act.io.read_arm_netcdf(result_ecor)
ds_ecorLoading...
# Create a plotting display object with 2 plots
display = act.plotting.TimeSeriesDisplay(ds_ecor, figsize=(15, 10))
co2_flux ='co2_flux'
ds_ecor = act.qc.add_dqr_to_qc(ds_ecor, variable=co2_flux)
display.plot(co2_flux, subplot_index=(0,))
# Add y-axis label with units (e.g., µmol CO₂ m⁻² s⁻¹)
display.axes[0].set_ylabel('CO₂ Flux (µmol m⁻² s⁻¹)')---------------------------------------------------------------------------
TimeoutError Traceback (most recent call last)
File /opt/conda/lib/python3.11/site-packages/urllib3/connection.py:174, in HTTPConnection._new_conn(self)
173 try:
--> 174 conn = connection.create_connection(
175 (self._dns_host, self.port), self.timeout, **extra_kw
176 )
178 except SocketTimeout:
File /opt/conda/lib/python3.11/site-packages/urllib3/util/connection.py:95, in create_connection(address, timeout, source_address, socket_options)
94 if err is not None:
---> 95 raise err
97 raise socket.error("getaddrinfo returns an empty list")
File /opt/conda/lib/python3.11/site-packages/urllib3/util/connection.py:85, in create_connection(address, timeout, source_address, socket_options)
84 sock.bind(source_address)
---> 85 sock.connect(sa)
86 return sock
TimeoutError: [Errno 110] Connection timed out
During handling of the above exception, another exception occurred:
ConnectTimeoutError Traceback (most recent call last)
File /opt/conda/lib/python3.11/site-packages/urllib3/connectionpool.py:715, in HTTPConnectionPool.urlopen(self, method, url, body, headers, retries, redirect, assert_same_host, timeout, pool_timeout, release_conn, chunked, body_pos, **response_kw)
714 # Make the request on the httplib connection object.
--> 715 httplib_response = self._make_request(
716 conn,
717 method,
718 url,
719 timeout=timeout_obj,
720 body=body,
721 headers=headers,
722 chunked=chunked,
723 )
725 # If we're going to release the connection in ``finally:``, then
726 # the response doesn't need to know about the connection. Otherwise
727 # it will also try to release it and we'll have a double-release
728 # mess.
File /opt/conda/lib/python3.11/site-packages/urllib3/connectionpool.py:404, in HTTPConnectionPool._make_request(self, conn, method, url, timeout, chunked, **httplib_request_kw)
403 try:
--> 404 self._validate_conn(conn)
405 except (SocketTimeout, BaseSSLError) as e:
406 # Py2 raises this as a BaseSSLError, Py3 raises it as socket timeout.
File /opt/conda/lib/python3.11/site-packages/urllib3/connectionpool.py:1060, in HTTPSConnectionPool._validate_conn(self, conn)
1059 if not getattr(conn, "sock", None): # AppEngine might not have `.sock`
-> 1060 conn.connect()
1062 if not conn.is_verified:
File /opt/conda/lib/python3.11/site-packages/urllib3/connection.py:363, in HTTPSConnection.connect(self)
361 def connect(self):
362 # Add certificate verification
--> 363 self.sock = conn = self._new_conn()
364 hostname = self.host
File /opt/conda/lib/python3.11/site-packages/urllib3/connection.py:179, in HTTPConnection._new_conn(self)
178 except SocketTimeout:
--> 179 raise ConnectTimeoutError(
180 self,
181 "Connection to %s timed out. (connect timeout=%s)"
182 % (self.host, self.timeout),
183 )
185 except SocketError as e:
ConnectTimeoutError: (<urllib3.connection.HTTPSConnection object at 0x7f505ba41fd0>, 'Connection to dqr-web-service.svcs.arm.gov timed out. (connect timeout=None)')
During handling of the above exception, another exception occurred:
MaxRetryError Traceback (most recent call last)
File /opt/conda/lib/python3.11/site-packages/requests/adapters.py:486, in HTTPAdapter.send(self, request, stream, timeout, verify, cert, proxies)
485 try:
--> 486 resp = conn.urlopen(
487 method=request.method,
488 url=url,
489 body=request.body,
490 headers=request.headers,
491 redirect=False,
492 assert_same_host=False,
493 preload_content=False,
494 decode_content=False,
495 retries=self.max_retries,
496 timeout=timeout,
497 chunked=chunked,
498 )
500 except (ProtocolError, OSError) as err:
File /opt/conda/lib/python3.11/site-packages/urllib3/connectionpool.py:801, in HTTPConnectionPool.urlopen(self, method, url, body, headers, retries, redirect, assert_same_host, timeout, pool_timeout, release_conn, chunked, body_pos, **response_kw)
799 e = ProtocolError("Connection aborted.", e)
--> 801 retries = retries.increment(
802 method, url, error=e, _pool=self, _stacktrace=sys.exc_info()[2]
803 )
804 retries.sleep()
File /opt/conda/lib/python3.11/site-packages/urllib3/util/retry.py:594, in Retry.increment(self, method, url, response, error, _pool, _stacktrace)
593 if new_retry.is_exhausted():
--> 594 raise MaxRetryError(_pool, url, error or ResponseError(cause))
596 log.debug("Incremented Retry for (url='%s'): %r", url, new_retry)
MaxRetryError: HTTPSConnectionPool(host='dqr-web-service.svcs.arm.gov', port=443): Max retries exceeded with url: /dqr_full/bnfecorsfS20.b1/20250401/20250430/incorrect,suspect (Caused by ConnectTimeoutError(<urllib3.connection.HTTPSConnection object at 0x7f505ba41fd0>, 'Connection to dqr-web-service.svcs.arm.gov timed out. (connect timeout=None)'))
During handling of the above exception, another exception occurred:
ConnectTimeout Traceback (most recent call last)
Cell In[30], line 4
2 display = act.plotting.TimeSeriesDisplay(ds_ecor, figsize=(15, 10))
3 co2_flux ='co2_flux'
----> 4 ds_ecor = act.qc.add_dqr_to_qc(ds_ecor, variable=co2_flux)
5 display.plot(co2_flux, subplot_index=(0,))
6 # Add y-axis label with units (e.g., µmol CO₂ m⁻² s⁻¹)
File /opt/conda/lib/python3.11/site-packages/act/qc/arm.py:155, in add_dqr_to_qc(ds, variable, assessment, exclude, include, normalize_assessment, cleanup_qc, dqr_link, skip_location_vars, create_missing_qc_variables)
152 url += f"/{assessment}"
154 # Call web service
--> 155 req = requests.get(url)
157 # Check status values and raise error if not successful
158 status = req.status_code
File /opt/conda/lib/python3.11/site-packages/requests/api.py:73, in get(url, params, **kwargs)
62 def get(url, params=None, **kwargs):
63 r"""Sends a GET request.
64
65 :param url: URL for the new :class:`Request` object.
(...)
70 :rtype: requests.Response
71 """
---> 73 return request("get", url, params=params, **kwargs)
File /opt/conda/lib/python3.11/site-packages/requests/api.py:59, in request(method, url, **kwargs)
55 # By using the 'with' statement we are sure the session is closed, thus we
56 # avoid leaving sockets open which can trigger a ResourceWarning in some
57 # cases, and look like a memory leak in others.
58 with sessions.Session() as session:
---> 59 return session.request(method=method, url=url, **kwargs)
File /opt/conda/lib/python3.11/site-packages/requests/sessions.py:589, in Session.request(self, method, url, params, data, headers, cookies, files, auth, timeout, allow_redirects, proxies, hooks, stream, verify, cert, json)
584 send_kwargs = {
585 "timeout": timeout,
586 "allow_redirects": allow_redirects,
587 }
588 send_kwargs.update(settings)
--> 589 resp = self.send(prep, **send_kwargs)
591 return resp
File /opt/conda/lib/python3.11/site-packages/requests/sessions.py:703, in Session.send(self, request, **kwargs)
700 start = preferred_clock()
702 # Send the request
--> 703 r = adapter.send(request, **kwargs)
705 # Total elapsed time of the request (approximately)
706 elapsed = preferred_clock() - start
File /opt/conda/lib/python3.11/site-packages/requests/adapters.py:507, in HTTPAdapter.send(self, request, stream, timeout, verify, cert, proxies)
504 if isinstance(e.reason, ConnectTimeoutError):
505 # TODO: Remove this in 3.0.0: see #2811
506 if not isinstance(e.reason, NewConnectionError):
--> 507 raise ConnectTimeout(e, request=request)
509 if isinstance(e.reason, ResponseError):
510 raise RetryError(e, request=request)
ConnectTimeout: HTTPSConnectionPool(host='dqr-web-service.svcs.arm.gov', port=443): Max retries exceeded with url: /dqr_full/bnfecorsfS20.b1/20250401/20250430/incorrect,suspect (Caused by ConnectTimeoutError(<urllib3.connection.HTTPSConnection object at 0x7f505ba41fd0>, 'Connection to dqr-web-service.svcs.arm.gov timed out. (connect timeout=None)'))
#display = act.plotting.TimeSeriesDisplay(ds_ecor, figsize=(15, 10))
#wind_direction_from_north ='wind_direction_from_north'
#ds_ecor = act.qc.add_dqr_to_qc(ds_ecor, variable=co2_flux)
#display.plot(wind_direction_from_north)
#rose_plot
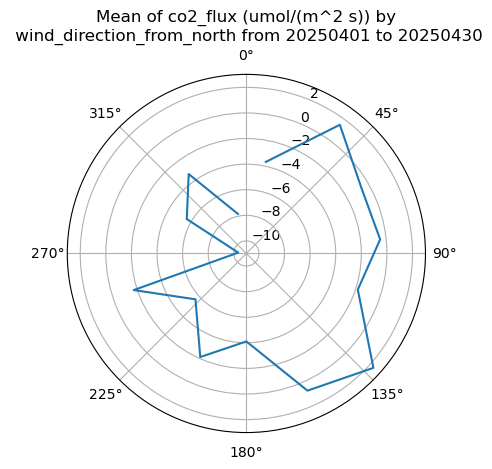
display = act.plotting.WindRoseDisplay(ds_ecor)
display.plot_data('wind_direction_from_north', 'mean_wind', 'co2_flux', num_dirs=15, plot_type='line', line_plot_calc='mean')
plt.show()
/opt/conda/lib/python3.11/site-packages/act/plotting/windrosedisplay.py:358: RuntimeWarning: Mean of empty slice
arr.append(np.nanmean(data[idx]))
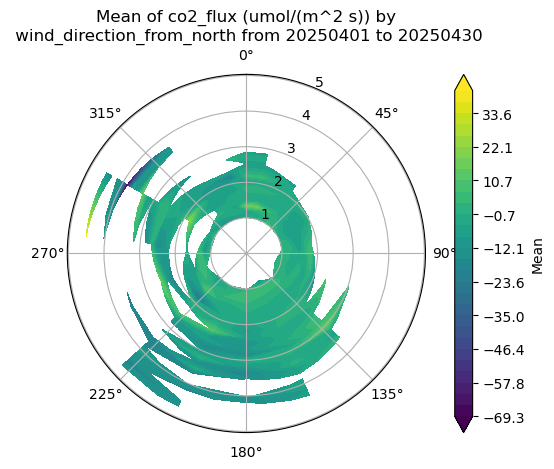
#rose_plot_contour
display = act.plotting.WindRoseDisplay(ds_ecor)
display.plot_data('wind_direction_from_north', 'mean_wind', 'co2_flux', num_dirs=15, plot_type='contour', contour_type='mean')
plt.show() /opt/conda/lib/python3.11/site-packages/act/plotting/windrosedisplay.py:429: RuntimeWarning: Mean of empty slice
mean_data[i, j] = np.nanmean(data[idx])
display = act.plotting.TimeSeriesDisplay(ds_ecor, figsize=(15, 10))
qc_co2_flux ='qc_co2_flux'
display.plot(qc_co2_flux, subplot_index=(0,))
<Axes: title={'center': 'bnfecorsfS20.b1 qc_co2_flux on 20250401'}, xlabel='Time [UTC]', ylabel='(1)'>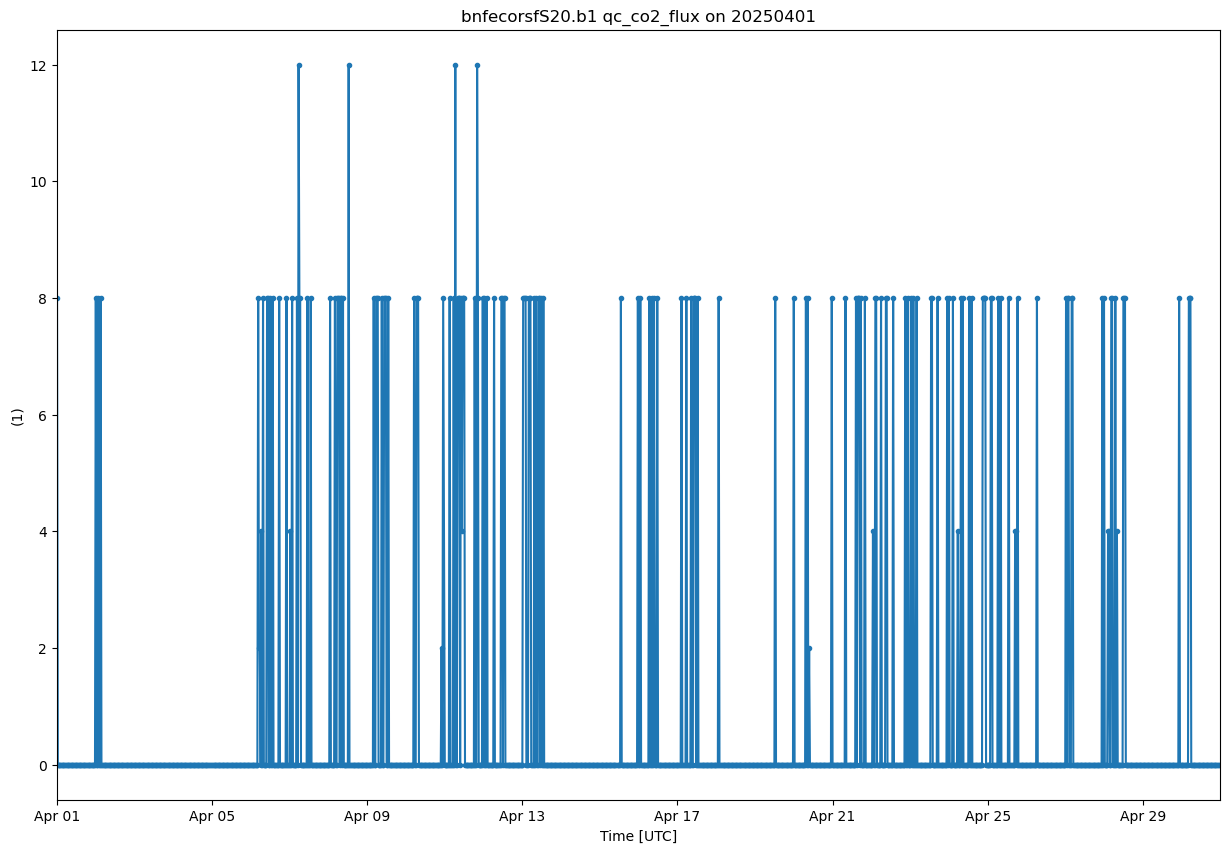
display = act.plotting.TimeSeriesDisplay(ds_ecor, figsize=(15, 10))
sensible_heat_flux ='sensible_heat_flux'
#ds_ecor = act.qc.add_dqr_to_qc(ds_ecor, variable=sensible_heat_flux)
display.plot(sensible_heat_flux)<Axes: title={'center': 'bnfecorsfS20.b1 sensible_heat_flux on 20250430'}, xlabel='Time [UTC]', ylabel='(W/m^2)'>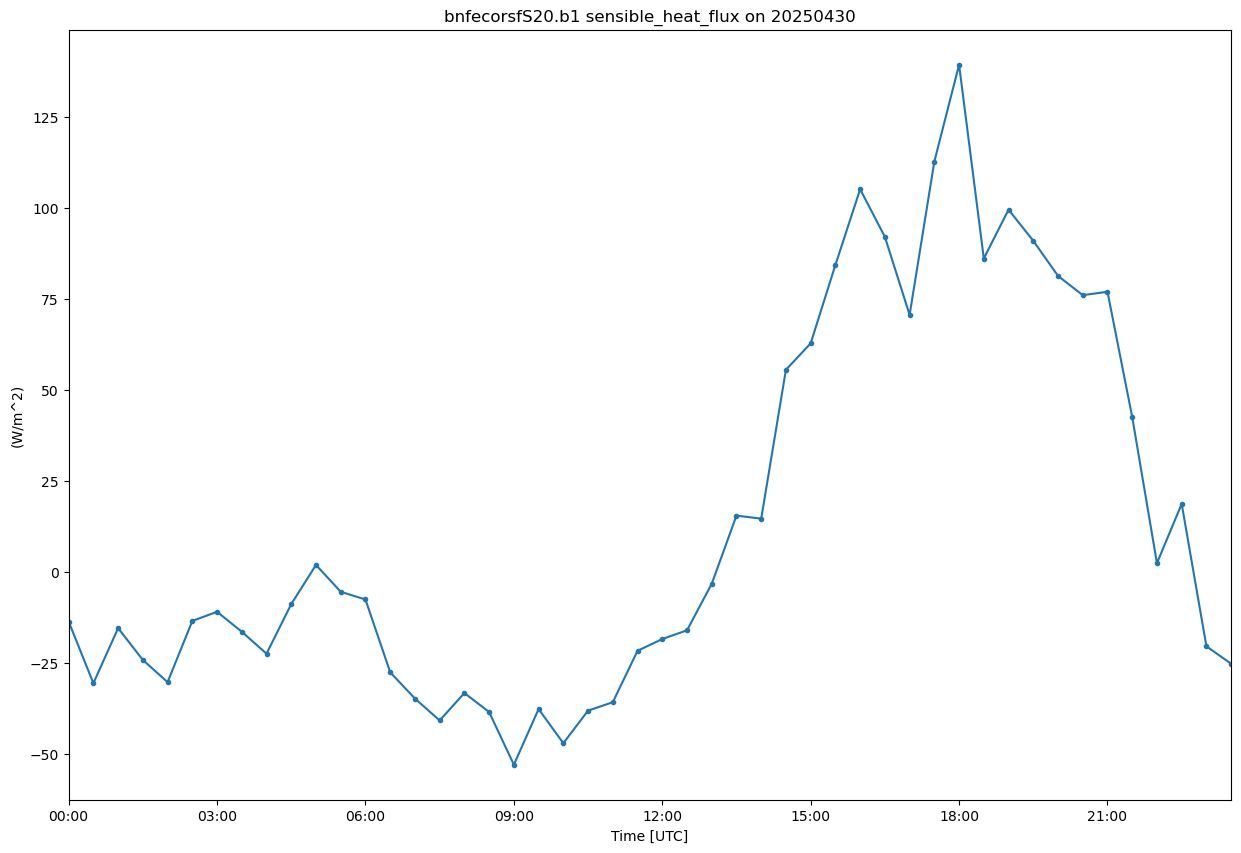
#Rn −G−S=H+LE (Rn= Net radiation (Total incoming - outgoing radiation at the surface), G = Ground heat flux, H= Sensible Heat Flux (Air), LE: Evapotranspiration Surface energy balance closure)